Lotus Base
Your browser is unable to support new features implemented in HTML5 and CSS3 to render this site as intended. Your experience may suffer from functionality degradation but the site should remain usable. We strongly recommend the latest version of Google Chrome, OS X Safari or Mozilla Firefox. As Safari is bundled with OS X, if you are unable to upgrade to a newer version of OS X, we recommend using an open source browser. Dismiss message
An integrated information portal for Lotus
Lotus Base is an integrated information portal for the model legume Lotus japonicus. Similar to the creation of online portals for other model plants, Lotus Base is motivated by the fragmented landscape of Lotus data, and strives to provide comprehensive data and a unified workflow to legume researchers. In the spirit of open research, Lotus Base is open source and free-of-charge.
Further information on how Lotus Base is implemented can be gleaned from our published paper:
Mun, T., Bachmann, A., Gupta, V., Stougaard, J., Andersen, S.U. (2016). Lotus Base: An integrated information portal for the model legume Lotus japonicus. Sci. Rep. doi:10.1038/srep39447.
Search
Looking for something? Use the search form below to start mining through currently available Lotus data.
Using LORE1 lines
The LORE1 lines are currently shipped free of charge and MTAs are not required. If you have used published protocols in establishing an independent LORE1 mutagenesis population, please cite the following papers, published back-to-back in Plant Journal:
- Urbanski et al. (2012). Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J., 69(4). doi:10.1111/j.1365-313X.2011.04827.x.
- Fukai et al. (2012) Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1 (2012). Plant J., 69(4). doi:10.1111/j.1365-313X.2011.04826.x.
For the general use of LORE1 mutants, we ask you to cite:
- Małolepszy et al. (2016). The LORE1 insertion mutant resource. Plant J. doi:10.1111/tpj.13243.
The majority of the LORE1 lines are released pre-publication, and the Centre for Carbohydrate Recognition and Signalling reserves the right to undertake and publish large-scale analysis of the insertion site data. Large-scale in this context refers to any sequence intervals or combinations thereof that exceed one megabase in length.
Should you encounter any difficulty in using this site, you can look up the end-user documentation.
Fresh from the blog
-
20 Apr 2020 L. japonicus Gifu v1.2 genome
L. japonicus Gifu v1.2 genome has been publicly released. It is now accessible via the Genome Browser, as well as other tools on the sites, such as the Expression Atlas (ExpAt) and View tools. -
21 Nov 2018 Post-mortem on downtime experienced by BLAST-related tools
-
12 Dec 2016 GateKeeper—Migrating away from IP-based controlled access
-

28 Nov 2016 Using InterProScan like a pro
Biologists are often challenged with this question when working with proteins. This article enunciates the process of using InterPro(Scan) to predict protein domain and function. Predicting protein domain(s) and therefore... -
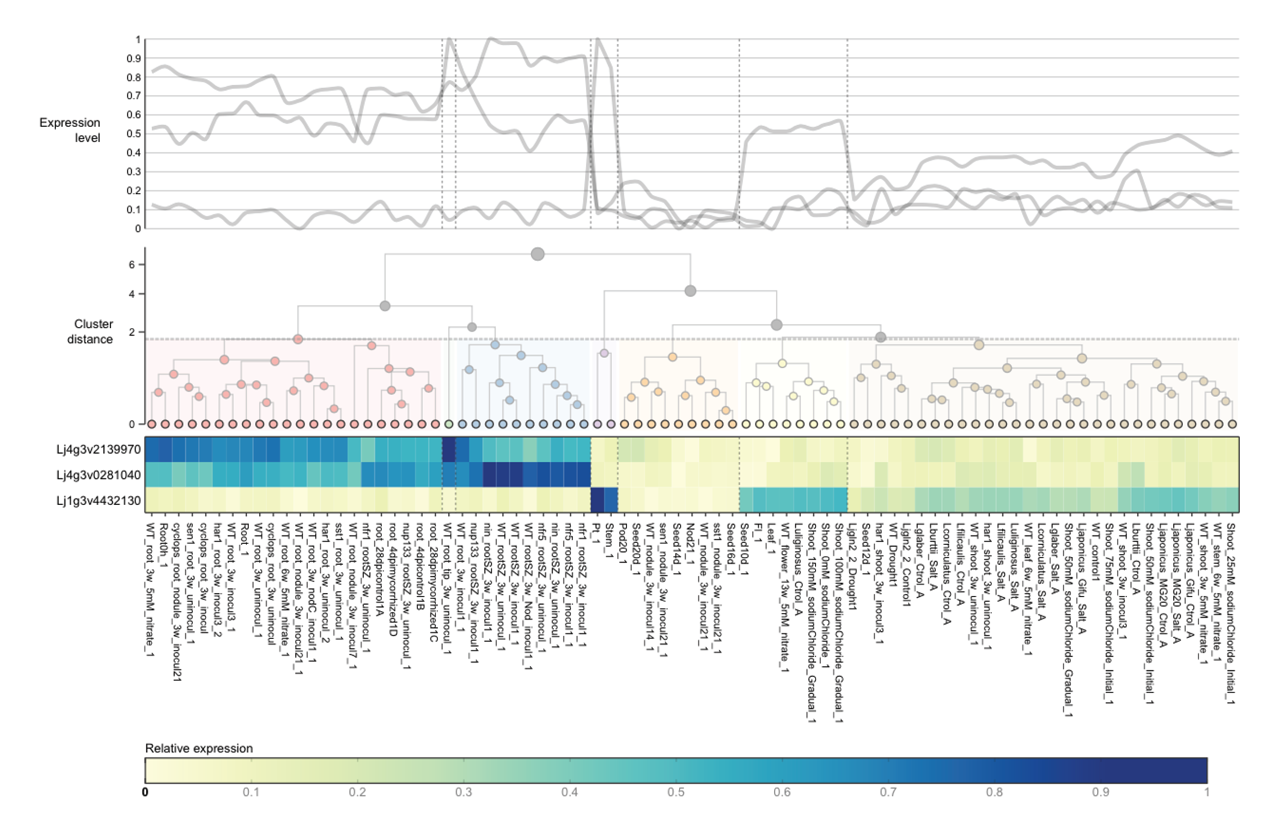
26 Aug 2016 Introducing ExpAt, the Lotus japonicus Expression Atlas
-
24 Aug 2016 Mapping transcripts across Lotus genome versions
-

05 Mar 2016 Redesigned LORE1 search form, and pan-version TREX searches
-
01 Mar 2016 JBrowse updated to v1.12.0
We have successfully upgraded our JBrowse installation, used to power visualization of the L. japonicus genome, to version 1.12.0.
Don't want to be left behind? Subscribe to our RSS feed to stay updated.
LORE1 mutants usage statistics
The provision of LORE1 insertional mutagenesis lines free-of-charge has allowed us to attract plant researchers across the globe to expand our current understanding and knowledge of the model legume Lotus japonicus. You can order your line(s) of interest now with our online form.

 Terry Mun
Terry Mun