Expression data from the model legume Lotus japonicus, while publicly available through other online resources, face a fragmented landscape that lacks accessibility and options for analysis and visualisation. Here we introduce ExpAt, the L. japonicus Expression Atlas, that offers features that empower legume researchers without the need for extensive knowledge in computation or skills in data visualisation.
What does ExpAt do?
ExpAt is a tool that allows you to query for the expression levels of your genes/transcripts of interest. It generates almost publication-ready, vector-based graphics based on the retrieved expression data, and presents them in a line graph and a heatmap. The line graph feature will be turned off when too many genes/transcripts were used in a single search, as it offers little insight on the expression patterns. You may export all the relevant data and charts by visiting the “export data” options that appears above the charts.
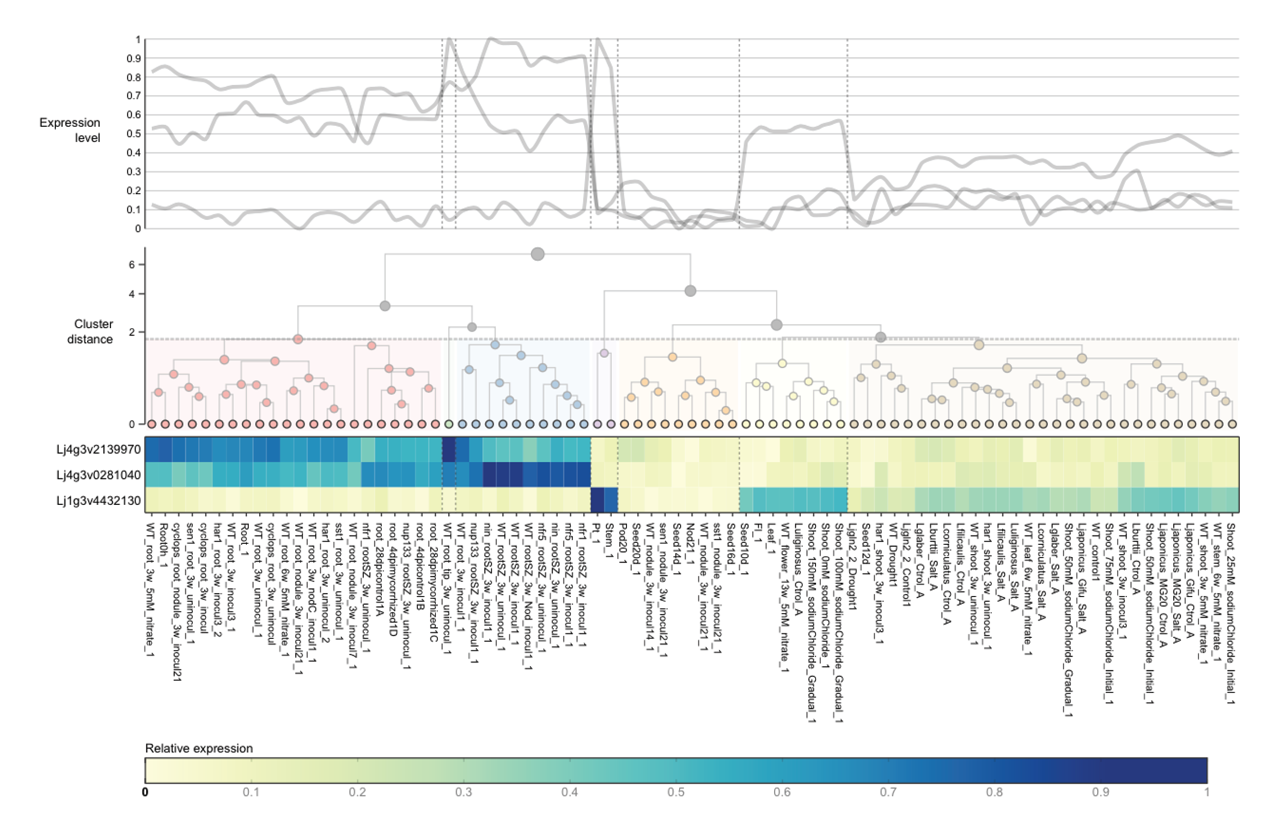
Here is an example of an unmodified ExpAt chart, showing a line graph and a clustered heatmap with one dendrogram on each axis:
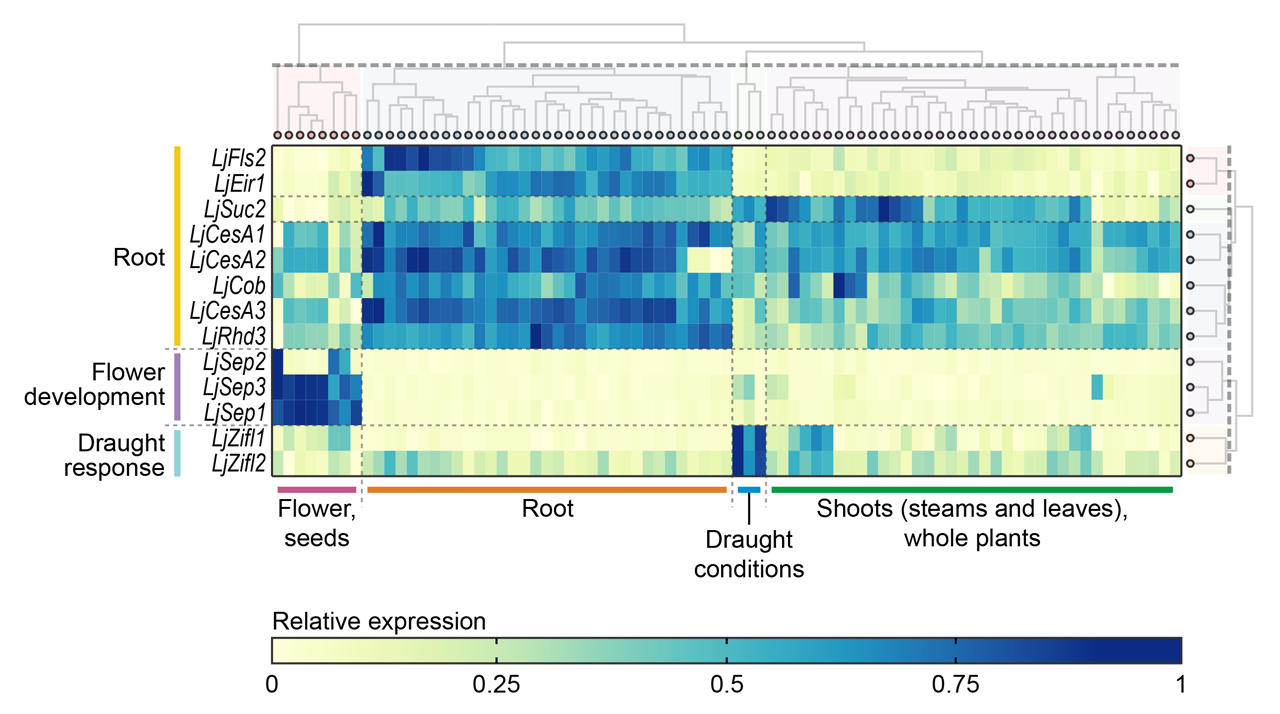
Here is an example of a ExpAt chart which is slightly tweaked in Adobe Illustrator, and used in a publication (Mun et al., in review):
Data transformation
For easing quick visual comparison across genes with signicantly different levels of absolute expression—measured by either (1) reads per kilobase of transcript (RPKM) for RNAseq datasets, or (2) arbitrary Affymetrix units for Affymetrix MicroArray datasets—we included two possibilities to transform the expression levels, by normalisation or standardisation.
Data normalisation is simply the rescaling of expression values to the domain $[0, 1]$, by subtracting the log-transformed sample expression levels $x_s$ with the lowest log-transformed expression level, $(\log_{10} x)_{\min}$, followed by the division of the difference between the log-transformed maximum and minimum expression levels. In order to allow comparison for extreme values, expression values are $\log_{10}$-transformed prior to normalisation.
Meanwhile, data standardisation serves to rescale the expression levels on a per row basis, across conditions, to have a mean of zero and a standard deviation of one. This is performed by subtracting the sample expression levels $x_s$ by the average expression level $\mu$ across all samples, and dividing the didderence with the sample standard deviation computed across all samples $\sigma$. This strategy is however erroneously labelled as “normalisation” in some studies.
Clustering
ExpAt offers the possibility to cluster your expression level data by gene/transcript idenifiers and/or conditions/samples. This two-dimensional data is referred to as a matrix—when this matrix has a dimension of $1 \times n$ or $n \times 1$, k-means clustering is used; when this matrix is larger than that, hierarchical agglomerative clustering is used. Changes to the clustering parameters can be modified on the fly and the heatmap and/or line graphs will be updated accordingly.
Dataset availability
We have integrated several publicly-available datasets:
- The L. japonicus gene expression (LjGEA)1–5, with probe identifiers mapped to L. japonicus v3.0 proteins using NCBI BLAST, and
- The early Lotus root responses to germinating spore exudates from arbuscular mycorrhizal fungi6.
Should you want to add your own expression data to the ExpAt tool, feel free to reach out to us via the contact form.
Citation
If you have used ExpAt for data transformation, analysis (k-means or hierarchical clustering), and/or visualisation, we ask that you cite Lotus Base7, and the relevant publications that generated the said dataset.
References
- Verdier, J., Torres-Jerez, I., Wang, M., Andriankaja, A., Allen, S. N., He, J., Tang, Y., Murray, J. D., and Udvardi, M. K. (2013). Establishment of the Lotus japonicus gene expression atlas (LjGEA) and its use to explore legume seed maturation. Plant J, 74(2):351–362.
- Díaz, P., Betti, M., Sánchez, D. H., Udvardi, M. K., Monza, J., and Márquez, A. J. (2010). De ciency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol, 188(4):1001–1013.
- Guether, M., Balestrini, R., Hannah, M., He, J., Udvardi, M. K., and Bonfante, P. (2009). Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol, 182(1):200–212.
- Høgslund, N., Radutoiu, S., Krusell, L., Voroshilova, V., Hannah, M. A., Go ard, N., Sanchez, D. H., Lippold, F., Ott, T., Sato, S., Tabata, S., Liboriussen, P., Lohmann, G. V., Schauser, L., Weiller, G. F., Udvardi, M. K., and Stougaard, J. (2009). Dissection of symbiosis and organ development by integrated transcrip- tome analysis of Lotus japonicus mutant and wild-type plants. PLoS One, 4(8):e6556.
- Sanchez, D. H., Lippold, F., Redestig, H., Hannah, M. A., Erban, A., Krämer, U., Kopka, J., and Udvardi, M. K. (2008). Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J, 53(6):973–987.
- Giovannetti, M., Mari, A., Novero, M., and Bonfante, P. (2015). Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front Plant Sci, 6:480.
- Mun, T., Bachmann, A., Gupta, V., Stougaard, J., and Andersen, S. U. (under review). Lotus base, an integrated information portal for Lotus japonicus.
