Tag “tools”
Lotus Base uses cookies to allow user preferences to be stored. By continuing to browse the site you are agreeing to our use of cookies.
Read more Accept cookie use and dismiss message
Your browser is unable to support new features implemented in HTML5 and CSS3 to render this site as intended. Your experience may suffer from functionality degradation but the site should remain usable. We strongly recommend the latest version of Google Chrome, OS X Safari or Mozilla Firefox. As Safari is bundled with OS X, if you are unable to upgrade to a newer version of OS X, we recommend using an open source browser. Dismiss message
-
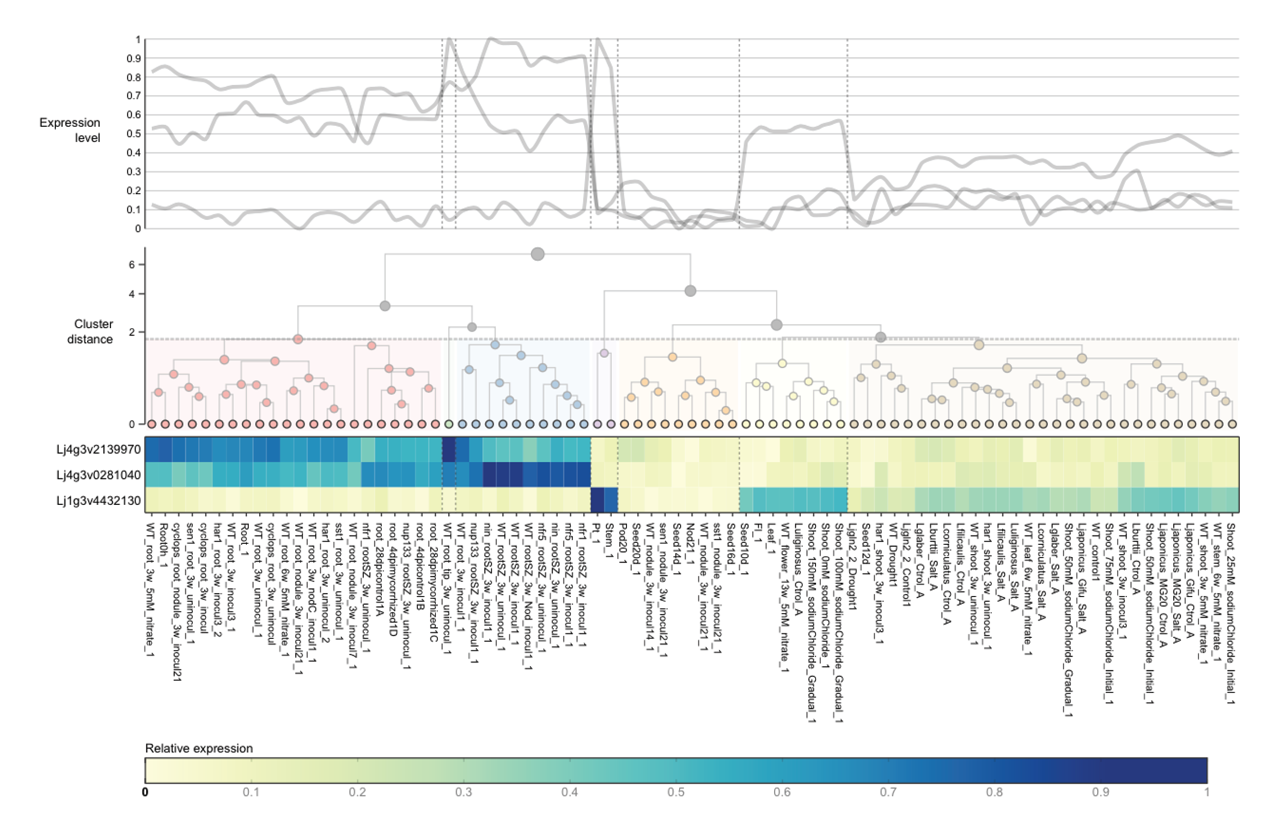
26 Aug 2016 Introducing ExpAt, the Lotus japonicus Expression Atlas
 Terry Mun
Terry Mun
- expat
- expression
- tools
Expression data from the model legume *Lotus japonicus*, while publicly available through other online resources, face a fragmented landscape that lacks accessibility and options for analysis and visualisation. Here we...
-
24 Aug 2016 Mapping transcripts across Lotus genome versions
 Terry Mun
Terry Mun
- tram
- tools
The availability of various versions of the *L. japonicus* genome, while proving to be an important resource in legume research, makes it difficult for users to map annotated genes and/or...
-

05 Mar 2016 Redesigned LORE1 search form, and pan-version TREX searches
 Terry Mun
Terry Mun
- lore1
- trex
- tools
*Lotus* Base was originally conceived as a very simple web interface for the searching for, and ordering of, LORE1 lines, but over the years it gradually evolved into a fully-fledged...
-
01 Mar 2016 JBrowse updated to v1.12.0
 Terry Mun
Terry Mun
- jbrowse
- tools
We have successfully upgraded our JBrowse installation, used to power visualization of the L. japonicus genome, to version 1.12.0.
-
22 May 2014 Retirement of LORE1 flanking sequences BLAST database
 Terry Mun
Terry Mun
- blast
- lore1
- tools
After much deliberation, we have decided to retire the LORE1 flanking sequences database because it does not contain LORE1 original reads, but only sequences that were extracted from the reference...
-
18 Nov 2013 New LORE1 lines available and BLAST update
 Terry Mun
Terry Mun
- lore1
- tools
- blast
LORE1 lines from batches 08, 09 and 10 are now available for searching. A new BLAST database has also been made available in light of the availability of new LORE1...
-
17 Oct 2013 TREX Tool
 Terry Mun
Terry Mun
- tools
- trex
The Transcript Explorer (TREX) tool is now available, and allows fetching of genome coordinates of Lotus transcripts based on their IDs.
-
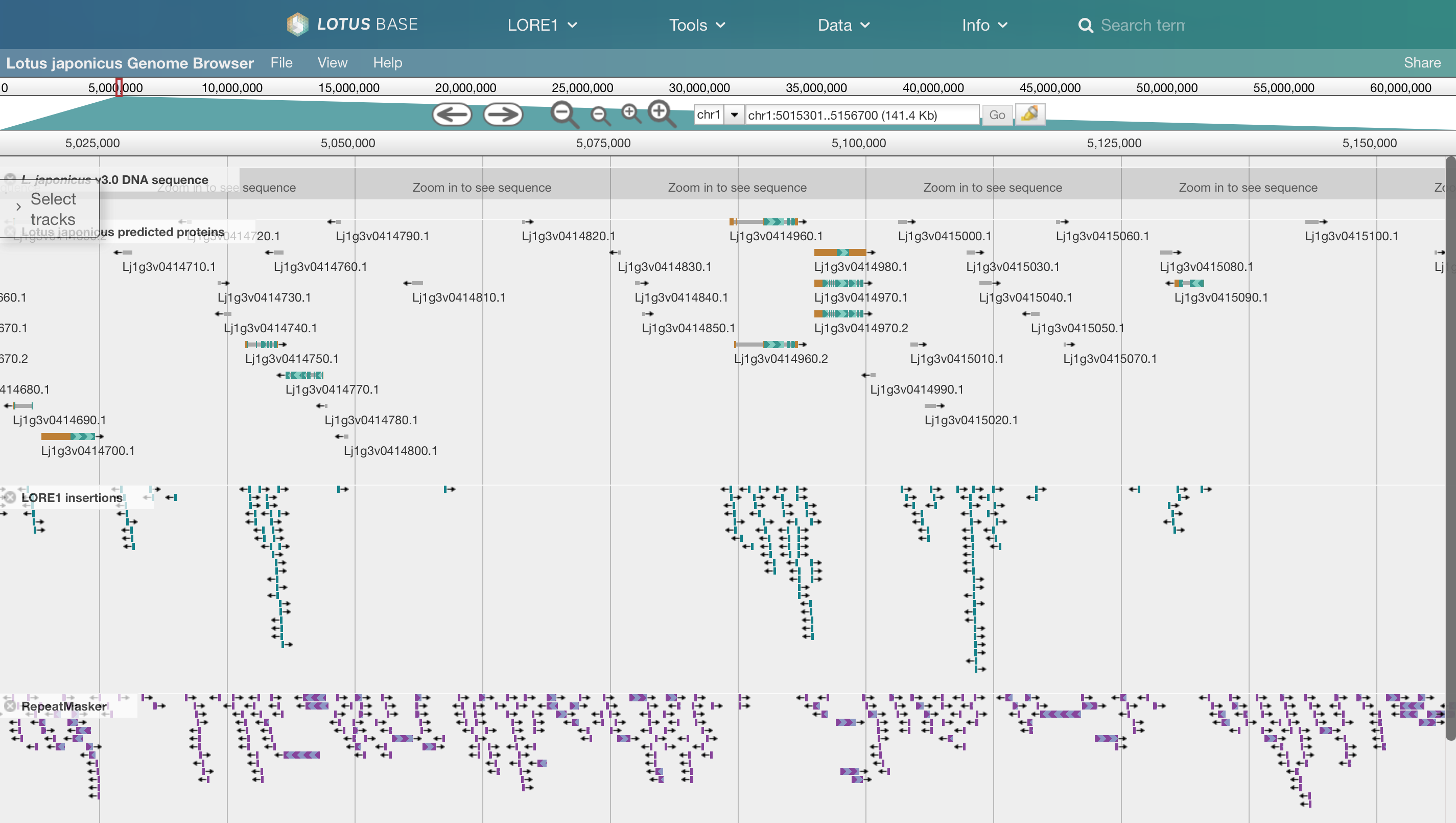
17 May 2013 Lotus genome browser
 Terry Mun
Terry Mun
- jbrowse
- tools
[](/genome) The [*Lotus* genome browser](/genome) is now ready to use. We will keep updating JBrowse to the latest version.
-
10 Apr 2013 Adjusting BLAST algorithm for short sequences
 Terry Mun
Terry Mun
- blast
- tools
If your BLAST queries are short (e.g. miRNA sequences), you might want to adjust the BLAST algorithm to get optimum results. Short sequences that are less than 20 bases will...