Biologists are often challenged with this question when working with proteins:
Now… what does your protein do?
Domain prediction—best friend or worst nightmare?
People want to know everything about your-favourite-protein-1 (YFP1). How does it look like? What are the predicted domains? Do these domains have any functions and processes associated with them? Are they located in specific parts of the cell?
A very simplified pipeline would be as follow:
- Check the amino acid sequence of YFP1 against various domain prediction programs
- Obtain domain and/or structural predictions of YFP1
- Infer biological function, molecular processes, and/or cellular components associated from said domain predictions
However, there are so many domain prediction algorithms out there, and an overwhelming bunch of them using Hidden Markov Models. These algorithms—such as PANTHER, Phobius, Pfam, SuperFamily, TMHMM—offer simple web interfaces that allows end-users to submit single (or a small number of, at best) sequences. EMBL-EBI offers InterPro, which integrates all of these prediction algorithms, but again only allows single sequence submission from their web interface.
There appears to be no simple way of submitting a set of protein sequence to multiple prediction algorithms—through a web interface, at least. If you are willing to dive into the world of command line interfaces, things start to look a bit better.
This article is written based on my experience with using InterPro, and my work with using RESTful services made available by EMBL-EBI on offering comprehensive Lotus data to legume researchers around the world.
Example use case: Lotus Base
As the principle developer and designer behind Lotus Base, I have worked on performing predictions on the entire set of predicted proteins using the most recently published Lotus japonicus genome — meaning 50,000+ predicted proteins in total that has to be parsed. The screenshot below shows an example of how I have pulled protein-specific data from a MySQL database built based on InterProScan’s domain prediction results, and merged the data with additional metadata obtained with the EB-eye REST service.
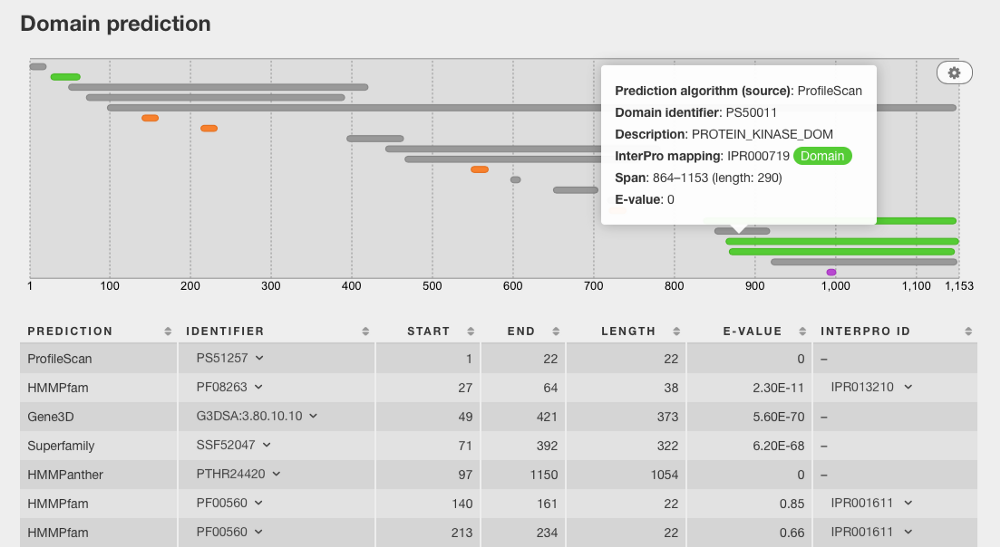
Of course, Lotus Base presents itself as a rather extreme use case due to the large volume of predicted proteins analysed. However, the methods described below would be just as applicable to a researcher who say, has obtained a list of proteins that are significantly enriched in one biological sample compared to another. The first step towards unraveling the functions of these proteins, based on their gene ontology predictions, would be to obtain their domain predictions first.
InterProScan vs InterPro RESTful service
You have two options from here on—if you are blessed with access to a computing cluster running on Linux, you can download and install a local version of InterProScan, and run InterProScan with FASTA files containing *n *number of sequences in parallel or in queue. The second, less handy option—but also the most accessible one—is to take advantage of the RESTful service provided by EMBL-EBI. The latter can be run on any computer, although preferably one running Unix/Linux (because that’s what my code will be running on). The only drawback is that EMBL-EBI’s fair use agreement only allows you to run InterProScan on 30 sequences at any one time.
Both services will give you the most up-to-date domain predictions, and necessitates re-running your proteins if they have included additional datasets. When InterProScan includes additional prediction algorithms, you can simply select to run said algorithms—instead of the entire set—on your sequences, and simply join the output with existing predictions.
Option A: Using EMBL-EBI’s InterPro REST service
Using the REST service provided by EMBL-EBI is a way to perform domain predictions on your protein(s) of interest without needing to invest in an expensive computing cluster, or obtaining access to one. For this part of the tutorial to work, you will need to ensure that Python3 is installed (InterPro provides a Python2 client library, but that is not covered in this section).
Explode FASTA file into individual sequence files
As the InterPro REST service only accepts single sequences, the easiest way is to split a multi-sequence FASTA file into individual sequence files. If your FASTA files are formatted such that each entry takes up two lines — one for the header and one for the sequence—you can do something like:
split -l 2 /path/to/your/fasta/file
However, this is often not the case, as FASTA files are recommended to be broken into lines containing no more than 60 characters long. If that is the case, you might want to rely on BioPython to do the parsing for you:
Hand individual FASTA file off to the REST service
When you have generated We can then iterate through these individual FASTA files and pass them to InterPro’s REST service. InterPro has provided us with various clients to interface with their REST service—I have chosen to work with their Python3 client. I did not modify their client script, with the exception of commenting out the line that prints the status in the function, so that my console will not be crowded with printouts.
It is important to respect the 30 sequences per batch limit of the InterPro REST service. Therefore, we will use a simple bash script that, while iterating through all individual FASTA file, stops after 30 files until the outcome from all 30 jobs have been returned:
If you want to obtain other output formats, remember to modify the option. According to my experience, each batch (of 30 sequences) takes around 2 minutes to complete.
The major drawback of this method is that it is a rather nuclear option if you are attempting to scan the entire collection of predicted proteins/transcripts. Use InterProScan on a computing cluster, if ever possible.
Option B: InterProScan on a computing cluster
Installing InterProScan
Follow the published instructions on installing InterProScan. I have ran into small hiccups, such as accidentally using an outdated version of Java (≤1.7) and having a dated GCC library. Loading the most updated one ensured that both InterProScan and the bundled BLAST+ package can be executed properly.
Adding proprietary algorithms
Note that InterProScan does not come with Phobius, SignalP, and TMHMM preinstalled. You will have to request for the compiled binaries of these algorithms, and upload them to their respective folders in the directory.
If you are unable to get hold of these libraries, you will have to retrieve the output of these algorithms via InterPro REST service.
A hitch with SignalP is that it assumes a fixed directory for loading the library files. This causes a fatal error where FASTA.pm cannot be loaded—remember to update the environment so that it points to the signalp directory (it will load libraries from the subfolder automagically).
# full path to the signalp-4.1 directory on your system (mandatory)
BEGIN {
$ENV{SIGNALP} = '<path/to/interproscan>/bin/signalp/4.1';
}
Check that all prediction algorithms are loaded
After you’ve done that, ensure that file is properly updated with the file paths of your binaries for the added libraries. After that, proceed to run without any arguments. It will print out all the algorithms that were detected and loaded correctly. Ensure that none is left behind—InterProScan will inform you if any of them has failed to load.
Depending on the number of sequences you want to submit per batch, you will have to update
Getting your FASTA files ready
You would want to process FASTA files in batches instead of all at one go. I have decided to split a unified FASTA file that contains all 50,000+ of the amino acid sequences into files containing 500 entries each. If your FASTA files are formatted such that each entry takes up two lines—one for the header and one for the sequence—you can do something like:
split -l 1000 /path/to/your/fasta/file
…assuming that you want 500 entries per file. However, this is often not the case, as FASTA files are recommended to be broken into lines containing no more than 60 characters long. If that is the case, you might want to rely on BioPython to do the parsing for you. The first step is to create a filtered FASTA file that is formatted such that each entry occupies two lines, generating a file. The second step is to batch parse this filtered file using itertools, to create batches of FASTA files containing 500 entries (i.e. 1000 lines) each:
Submit your jobs to iteratively to the computing cluster
In this case, I am using SLURM for batch job submission. I will not go into details on how the job submission is done, as it is highly dependent on the configuration of individual clusters. The actual command is quite simple:
/path/to/interproscan.sh \
-i /path/to/fasta.fa -dp -iprlookup --goterms --pathways
Note that I have turned off precalcualted match lookup using the flag because the computing cluster I am on blocks external connections for security reasons. Moreover, the *Lotus japonicus *proteins are yet to be submitted to UniProt so it is highly unlikely that we will find too many matching proteins in the public database.
Here is an example of how a batch job submission template you can use:
Boom! Run it and wait for magic to happen.
Performance
In the case of Lotus Base and our collection of predicted transcripts, we have 49,598 sequences scanned in batches of 1,000, creating 50 jobs. The jobs were run with an allocated 24Gb memory over 12 cores, on nodes equipped with Intel “Sandy Bridge” E5–2670 (2.67GHz)or “Haswell” E5–2680v3 (2.5GHz) CPUs. After normalizing for processor speeds and library sizes, the real time consumed per job stands at 2.50±0.28h (CPU time: 4.32±0.35h).
Parsing InterPro/InterProScan outputs
The file that contains all the juicy data is the TSV file, which you can easily import into a relational database such as MySQL. The InterProScan wiki has the information on what does each individual column in the TSV file contain.
For Lotus Base, I simply imported the TSV file *as-is *into a MySQL table, and used statements to merge transcript metadata from additional tables we have. It’s as simple as that!
This article is also published on Medium.com.
